-

-
Jeffrey E Barrick
Associate Professor
Molecular BiosciencesMicrobial experimental evolution and synthetic biologyjbarrick@cm.utexas.edu
Phone: 512-471-3247
Office Location
MBB 1.436
Postal Address
2500 SPEEDWAY
AUSTIN, TX 78712-
B.S., California Institute of Technology (2001)
Ph.D., Yale University (2006)
Postdoctoral Fellow, Michigan State University (2006-2010)Research
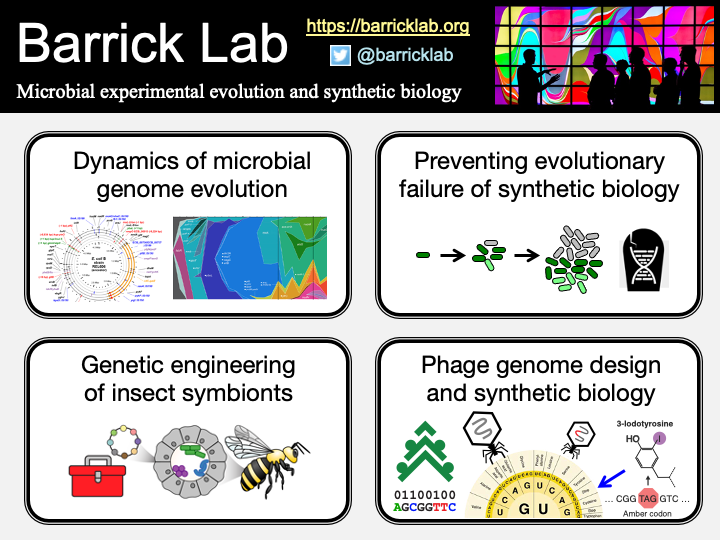
Controlling evolution is key to combating the emergence of drug resistance, preventing the progression of chronic infections and cancers, and maintaining the function of genetically engineered cells. Evolution can also be harnessed for biotechnology and medicine: used to create novel therapeutics and to optimize microbial bioprocessing, for example. My laboratory is interested in developing methods to anticipate and arrest unwanted evolution and in understanding how expanded genetic codes and other engineered genome-wide changes can be used to study and augment the evolutionary potential of microbes. Recently, we have also begun to use synthetic biology tools to engineer bacterial endosymbionts of insects for various applications, including engineering honey bee gut symbionts to protect pollinator health, engineering aphid symbionts to protect crops, and engineering leafhopper symbionts to control biomaterial production.
-
View all publications on barricklab.org
View all publications on Google Scholar
Representative Publications
Powell, J.E., Perutka, J., Geng, P., Heckmann, L.C., Horak, R.D., Davies, B.W., Ellington, A.D., Barrick, J.E., Moran, N.A. (2020) Engineered symbionts activate honey bee immunity and limit pathogens. Science 367:573–576.
Zhang, X., Deatherage, D.E., Zheng, H., Georgoulis, S.J., ^Barrick, J.E. (2019) Evolution of satellite plasmids can stabilize the maintenance of newly acquired accessory genes in bacteria. Nat. Comm. 10:5809.Leonard, S.P.,
Geng, P., Leonard, S.P., Mishler, D.M., Barrick, J.E. (2019) Synthetic genome defenses against selfish DNA elements stabilize engineered bacteria against evolutionary failure. ACS Synth. Biol. 8:521-531.
Deatherage, D.E., Leon, D., Rodriguez, Á.E., Omar, S., Barrick, J.E. (2018) Directed evolution of Escherichia coli with lower-than-natural plasmid mutation rates. Nucleic Acids Res. 46:9236-9250.
Leonard, S.P., Perutka, J., Powell, J.E., Geng, P., Richhart, D., Byrom, M., Kar, S., Davies, B.W., Ellington, A.D., Moran, N.A., Barrick, J.E. (2018) Genetic engineering of bee gut microbiome bacteria with a toolkit for modular assembly of broad-host-range plasmids. ACS Synth. Biol. 7:1279-1290.
Bull, J.J., Barrick, J.E. (2017) Arresting evolution. Trends Genet. 33:910-920.
Tenaillon, O., Barrick, J.E., Ribeck, N., Deatherage, D.E., Blanchard, J.L, Dasgupta, A., Wu, G.C., Wielgoss, S., Cruveiller, S., Medigue, C., Schneider, D., Lenski, R.E.^ (2016) Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 536:165-170.
Renda, B.A., Hammerling, M.J., Barrick, J.E. (2014) Engineering reduced evolutionary potential for synthetic biology. Mol. Biosyst. 10:1668-1678.
Deatherage, D.E., Barrick, J.E. (2014) Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151:165-188.
Hammerling, M.J., Ellefson, J.W., Boutz, D.R., Marcotte, E.M., Ellington, A.D., Barrick, J.E. (2014) Bacteriophages use an expanded genetic code on evolutionary paths to higher fitness. Nat. Chem. Biol. 10:178-180.
Barrick, J.E., Lenski, R.L. (2013) Genome dynamics during experimental evolution. Nat. Rev. Genet. 14:827-834.
-
- Defense Science Study Group (2018-2019)
- NSF CAREER Award (2016)
- NIH Pathway to Independence Award (2009)
- NSF Postdoctoral Research Fellowship in Biological Informatics (2006)
- RNA Society/Scaringe Young Scientist Award (2006)
- Howard Hughes Medical Institute Predoctoral Fellowship (2001)
- Caltech George W. Green Memorial Prize (2000)
-
BCH 339F: Foundations of Biochemistry
NSC 110: Biotechnology and Society
-



